Modern and Ancient Genomes
The Modern and Ancient Genomes group headed by Prof. Gabriel Renaud is found at the Université Laval. We are broadly interested in methods applied to next-generation sequencing data, ancient and environmental DNA, genotyping, allele frequencies and resolved haplotypes from either modern and ancient genomes as well as sedimentary DNA.

Highlights
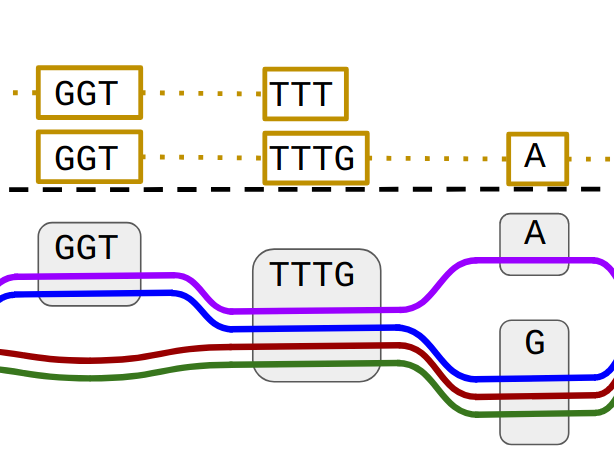
Pangenomes graph
Pangenome graphs are a data structure to represent the known diversity of a species instead of just using a linear reference. Several computational tools are being developed actively to build such graphs and subsequently align DNA sequencing reads to them. In our group, we make extensive use of pangenome graphs in combination with Bayesian inference.

Ancient DNA
Ancient DNA (aDNA) is degraded DNA extracted from fossils, museum samples etc. aDNA offers an amazing opportunity to study ancient species but is plagued by fragmentation, short fragment length, chemical damage and contamination. Our group develops novel computational methods to deal with such idiosyncrasies.

Sedimentary DNA
Sedimentary DNA (sedaDNA) is ancient environmental DNA that involves retrieving genetic material from environments where multiple organisms will contribute to it. However, the computational analysis of sedaDNA both faces the problems of ancient DNA and environmental DNA where multiple contributing species need to be deconvoluted. We craft exciting new tools to be able to analyze sedaDNA datasets for hominins, eukaryotes and microbes.
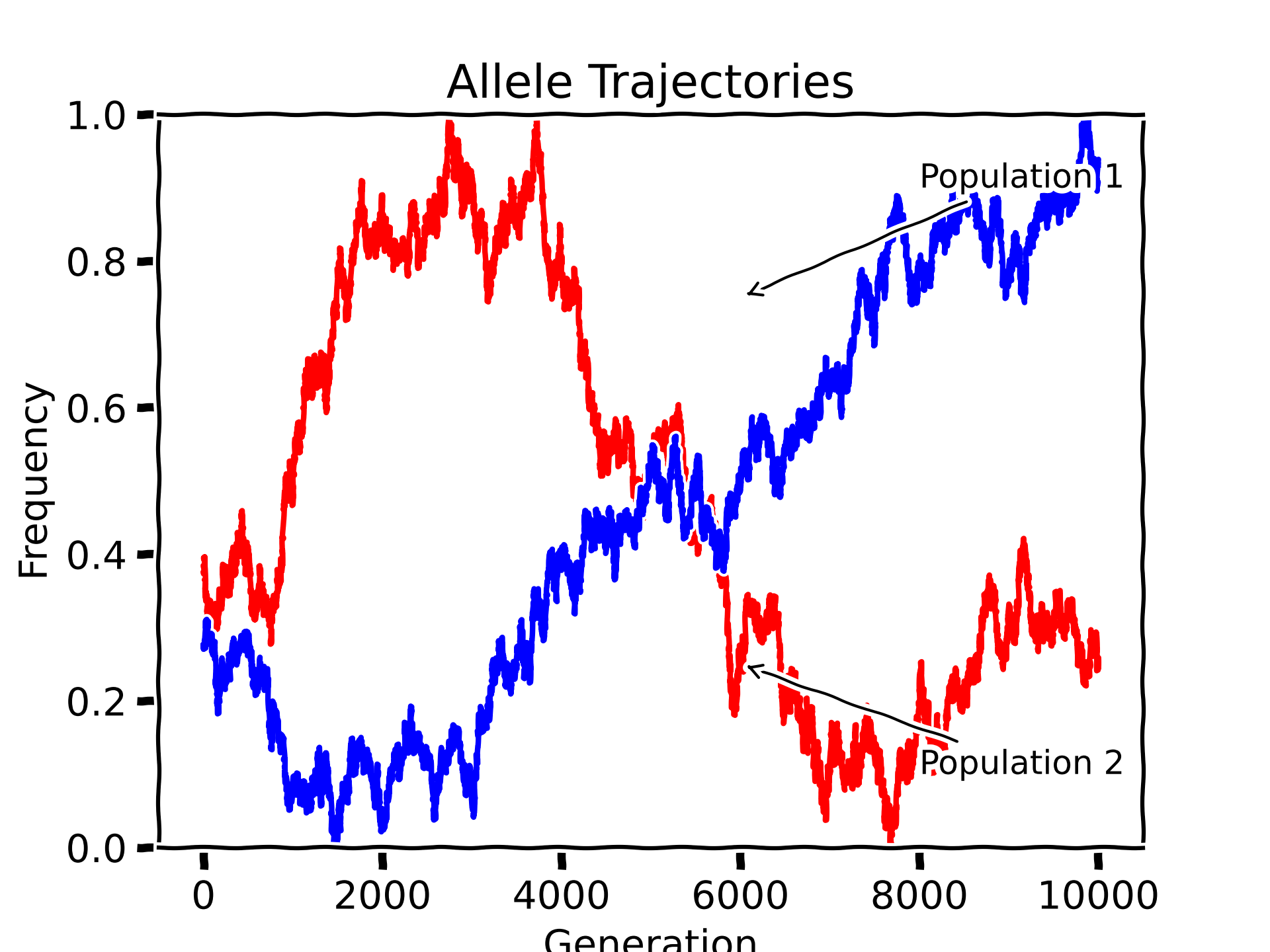
Allele frequencies
Our group develops computational methods to analyze, store and use ancient allele frequencies. By studying genetic diversity in ancient populations, we can gain insights into the history and evolution of various species including humans. Our methods utilize advanced algorithms and statistical models to analyze large-scale datasets from modern and ancient sources.
Our Tools
Science needs to be open, reproducible and accessible to all. This is why our developed tools and analyzed data are made public on GitHub and released under GPL or MIT licenses.
Our courses
Training the next generation of new scientists and bioinformaticians is crucial. We are responsible for 2 courses at DTU which are open to DTU students as well as students from abroad and people in industry.

Our Team
Our team consists of a diverse group of passionate scientists who possess unique interests and backgrounds. We all share a common goal of crafting novel computational methods for ancient and modern DNA as well as using existing tools to answer exciting scientific questions. To achieve this, we work closely with various other groups from different parts of the world that generate interesting empirical datasets.
Some images on this page were generated by DALL-E and Midjourney.
